Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
The actin filament plays a fundamental role in numerous cellular processes such as cell growth, proliferation, migration, division, and locomotion. The actin cytoskeleton is highly dynamical and can polymerize and depolymerize in a very short time under different stimuli. To study the mechanics of actin filament, quantifying the length and number of actin filaments in each time frame of microscopic images is fundamental. Different from microtubules, actin filaments is considered as from intersections/endpoints to intersection/endpoints, namely indiviudal segments of the actin network. In this project, we adopt a Convolutional Neural Network (CNN) to segment actin filaments first, and then we utilize a modified Resnet to detect junctions and endpoints of filaments. With binary segmentation and detected keypoints, we apply a fast marching algorithm to obtain the number and length of each actin filament in microscopic images. We have also collected a dataset of 10 microscopic images of actin filaments to test our method. Our experiments show that our approach outperforms other existing approaches tackling this problem regarding both accuracy and inference time. 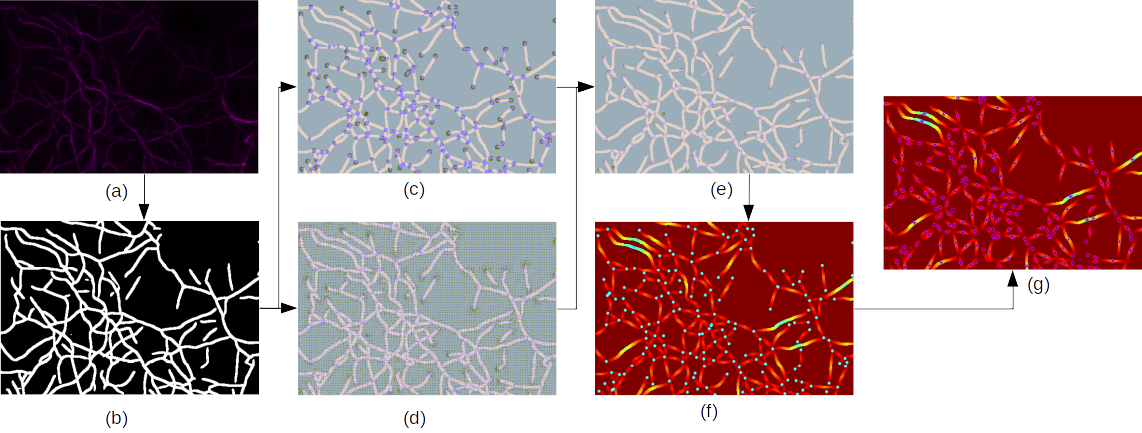
Filamentous structures play an important role in biolog-ical systems. Extracting individual filaments is fundamental for analyzing and quantifying related biological processes. However, segmenting filamentous structures at an instancelevel is hampered by their complex architecture, uniformappearance, and image quality. In this project, I propse an orientation-aware neural network, which contains six orientation-associated branches. Each branch detects filaments with specific range of orientations, thus separatingthem at junctions, and turning intersections to overpasses. A terminus pairing algorithm is also proposed to regroupfilaments from different branches, and achieve individual filaments extraction. 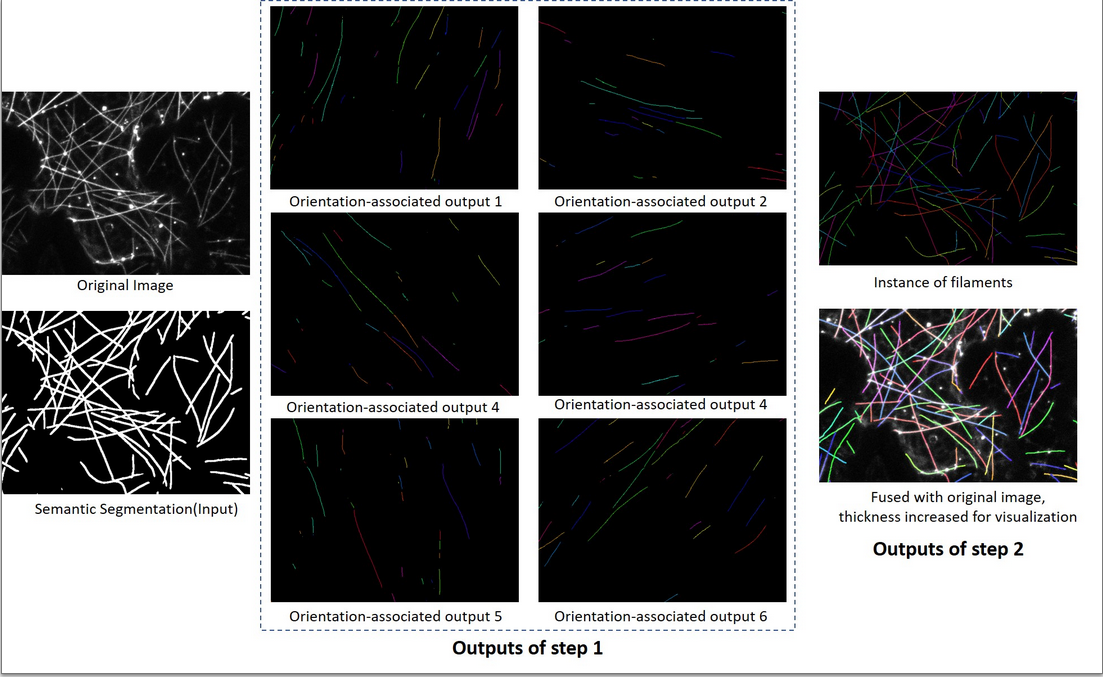
Collaborated with Ocean Engineering Lab at the University of Delaware and developed methods for automated quantification analysis of munition mobility experiments in a wave flume based on videos of a bird view camera 
Published in Journal 1, 2009
This paper is about the number 1. The number 2 is left for future work.
Recommended citation: Your Name, You. (2009). "Paper Title Number 1." Journal 1. 1(1). http://academicpages.github.io/files/paper1.pdf
Published in Journal 1, 2010
This paper is about the number 2. The number 3 is left for future work.
Recommended citation: Your Name, You. (2010). "Paper Title Number 2." Journal 1. 1(2). http://academicpages.github.io/files/paper2.pdf
Published in Journal 1, 2015
This paper is about the number 3. The number 4 is left for future work.
Recommended citation: Your Name, You. (2015). "Paper Title Number 3." Journal 1. 1(3). http://academicpages.github.io/files/paper3.pdf
Published:
This is a description of your talk, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Published:
This is a description of your conference proceedings talk, note the different field in type. You can put anything in this field.
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.